GASTON - Mapping the topography of spatial gene expression with interpretable deep learning
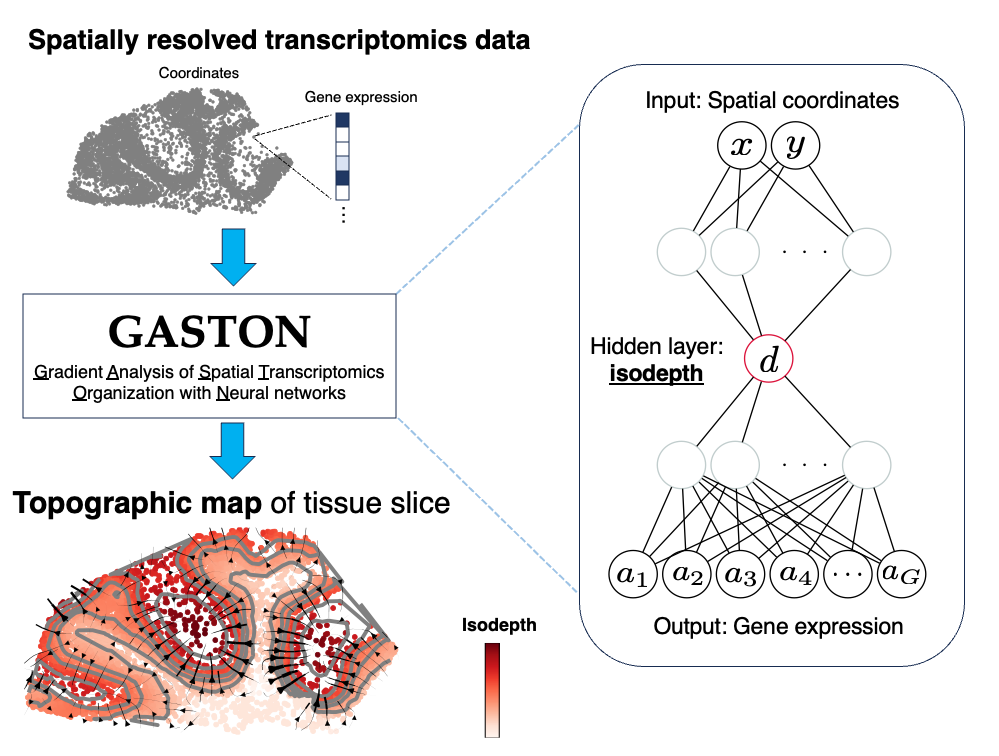
GASTON is an interpretable deep learning model for learning the gene expression topography of a tissue slice from spatially resolved transcriptomics (SRT) data. GASTON models gene expression topography by learning the isodepth, a 1-D coordinate describing continuous gene expression gradients and tissue geometry (i.e. spatial domains).
GASTON’s key applications
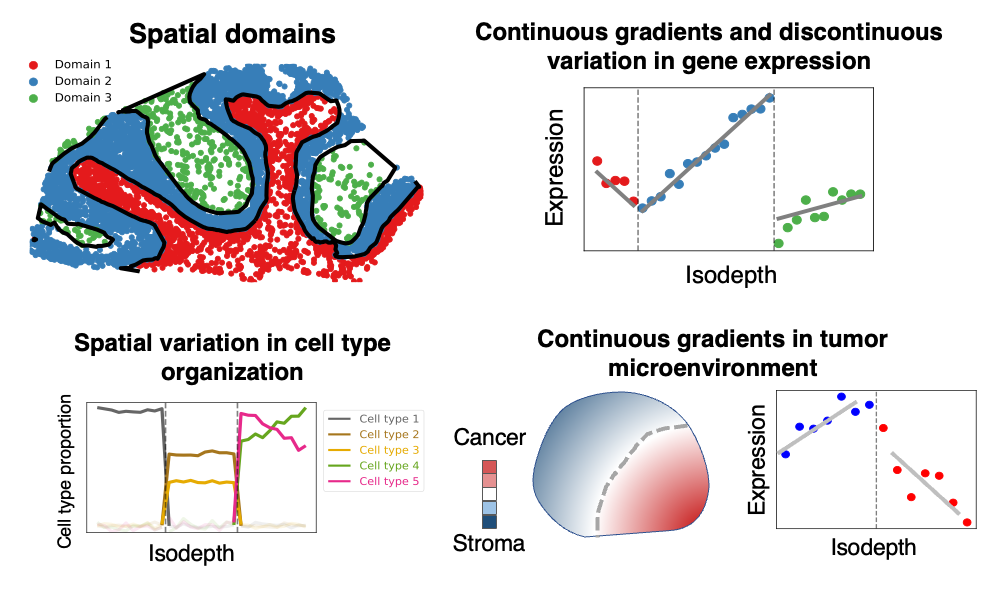
Learns 1-d coordinate that varies smoothly across tissue slice, providing topographic map of gene expression in the tissue slice.
Modeling continuous gradients of gene expression for individual genes, e.g. gradients of metabolism in cancer
Identifying tissue geometry, i.e. arrangement of spatial domains
Manuscript
Please see our manuscript [] for more details.